AI model can design billions of superbug-fighting antibiotic molecules
Mar. 22, 2024.
2 min. read
26 Interactions
Generative model can access tens of billions of promising molecules quickly and cheaply
Researchers at McMaster University and Stanford University have invented a new generative artificial intelligence model called SyntheMol that can design billions of new antibiotic molecules that are inexpensive and easy to build in the laboratory.
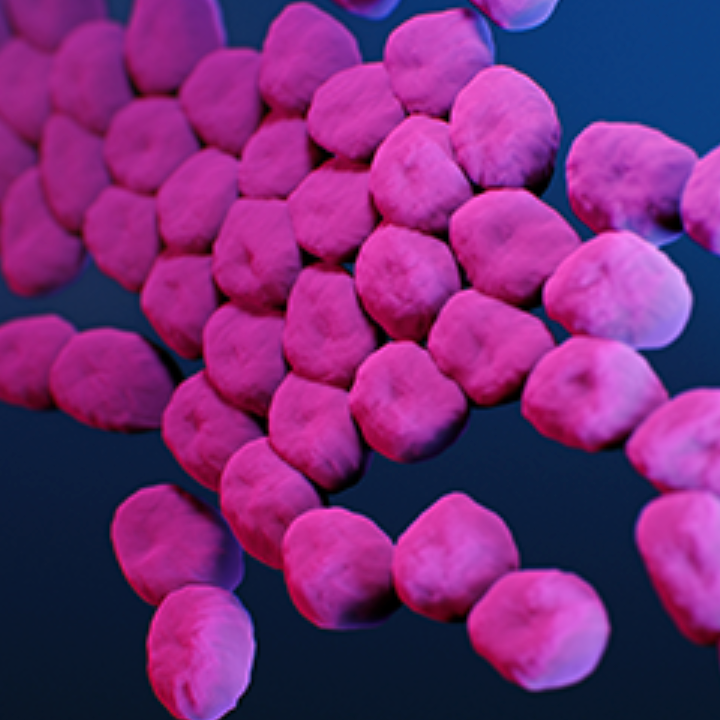
In a new study, published today in the journal Nature Machine Intelligence, researchers report that SyntheMol can design new antibiotics to stop the spread of Acinetobacter baumannii, which the World Health Organization has identified as one of the world’s most dangerous antibiotic-resistant bacteria.
A. baumannii can cause pneumonia and meningitis, and infect wounds, all of which can lead to death. Researchers say few treatment options remain.
Accessing tens of billions of promising molecules
Researchers developed the generative model to access tens of billions of promising molecules quickly and cheaply. They drew from a library of 132,000 molecular fragments, which fit together like Lego pieces but are all very different in nature.
They then cross-referenced these molecular fragments with a set of 13 chemical reactions, enabling them to identify 30 billion two-way combinations of fragments to design new molecules with the most promising antibacterial properties.
Zappjng world’s most dangerous antibiotic-resistant bacteria
Each of the molecules designed by this model was in turn fed through another AI model trained to predict toxicity. The process yielded six non-toxic molecules that display potent antibacterial activity against A. baumannii.
“Synthemol not only designs novel molecules that are promising drug candidates, but it also generates the recipe for how to make each new molecule. Generating such recipes is a new approach and a game changer because chemists do not know how to make AI-designed molecules,” says James Zou, an associate professor of biomedical data science at Stanford University, who co-authored the paper.
The research is funded in part by the Weston Family Foundation, the Canadian Institutes of Health Research, and Marnix and Mary Heersink.
Citation: Swanson, K., Liu, G., Catacutan, D.B. et al. Generative AI for designing and validating easily synthesizable and structurally novel antibiotics. Nat Mach Intell 6, 338–353 (2024). https://doi.org/10.1038/s42256-024-00809-7
Let us know your thoughts! Sign up for a Mindplex account now, join our Telegram, or follow us on Twitter.

.png)

.png)


.png)





7 Comments
7 thoughts on “AI model can design billions of superbug-fighting antibiotic molecules”
Amazing post,,
🟨 😴 😡 ❌ 🤮 💩
nice project
🟨 😴 😡 ❌ 🤮 💩
i like this post
🟨 😴 😡 ❌ 🤮 💩
I like it
🟨 😴 😡 ❌ 🤮 💩
interesting
🟨 😴 😡 ❌ 🤮 💩
Yes, it will be interesting to see how this plays out in the drug industry.
🟨 😴 😡 ❌ 🤮 💩
AI's low hanging fruits with huge benefits. Happy to see progress.
🟨 😴 😡 ❌ 🤮 💩