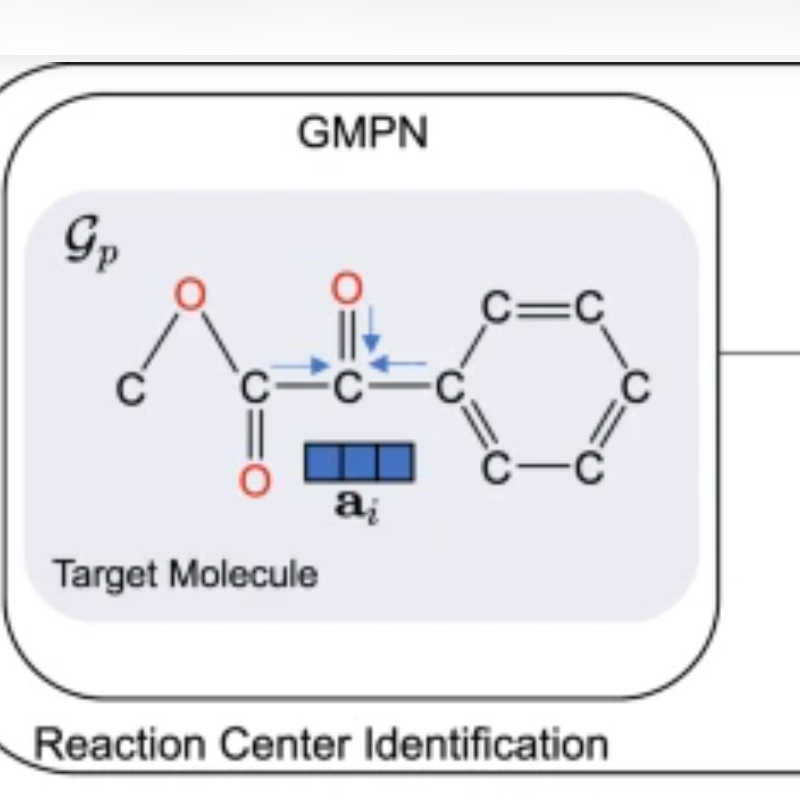
Researchers at The Ohio State University have created an AI framework called G2Retro that automatically generates reactions for any given molecule.
G2Retro covers an enormous range of possible chemical reactions and can accurately and quickly determine which reactions might work best.
The study was published May 30 in the journal Communications Chemistry.
The researchers trained G2Retro on a dataset that contains 40,000 chemical reactions collected between 1976 and 2016. The framework “learns” from graph-based representations of given molecules, and uses deep neural networks to generate possible reactant structures that could be used to synthesize them.
Hundreds of new reaction predictions
Given a molecule, G2Retro can come up with hundreds of new reaction predictions in only a few minutes, said Xia Ning, PhD, lead author of the study, an associate professor of computer science and engineering at Ohio State and an associate professor of biomedical informatics in the College of Medicine.
“G2Retro is able to supply multiple synthesis routes and options, as well as a way to rank different options for each molecule,” said Dr. Ning. “This is not going to replace current lab-based experiments, but it will offer more and better drug options so experiments can be prioritized and focused much faster.”
This research was supported by the National Science Foundation and Ohio State’s President’s Research Excellence Program.
Citation: Chen, Z., Ayinde, O. R., Fuchs, J. R., Sun, H., & Ning, X. (2023). G2Retro as a two-step graph generative models for retrosynthesis prediction. Communications Chemistry, 6(1), 1-19. https://doi.org/10.1038/s42004-023-00897-3 (open-access).
Let us know your thoughts! Sign up for a Mindplex account now, join our Telegram, or follow us on Twitter.

.png)

.png)


.png)