Deep learning system for protein design
Dec. 16, 2024.
2 mins. read.
1 Interactions
Researchers have developed a new, efficient AI tool that uses deep learning to design proteins that can bind to small molecules.
Researchers from the University of Science and Technology of China and Harvard Medical School have developed a new artificial intelligence (AI) tool called PocketGen.
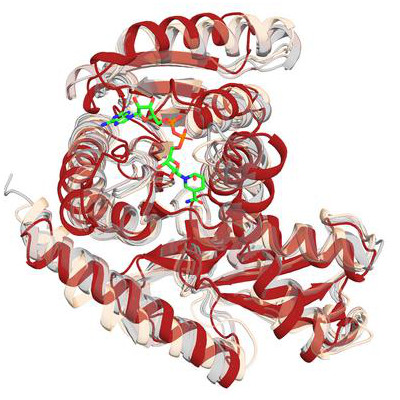
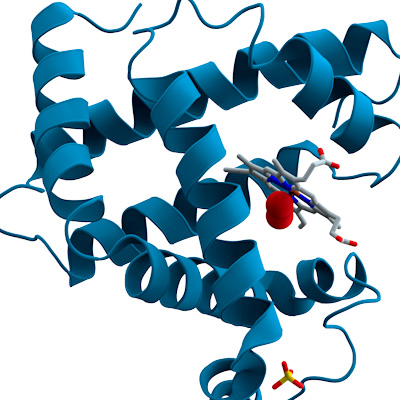
Pockets are specific regions on the surface of a protein where small molecules, like drugs or ligands, can bind.
PocketGen uses deep learning to design proteins that can bind to small molecules. Binding means the protein and molecule stick together in a useful way. This can help make new medicines or sensors.
Traditional ways to design these proteins are slow and often don’t work well. They use energy optimization, which tries to find the best shape for a protein, and template matching, where they copy the shape of known proteins. These methods struggle because they don’t handle the complexity of how proteins and molecules interact.
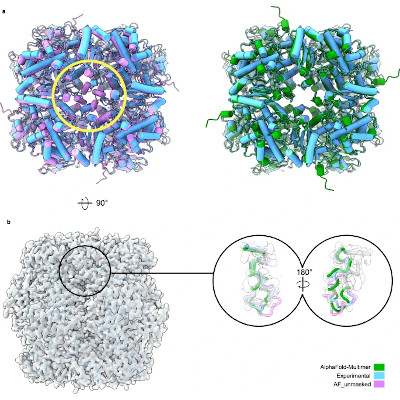
PocketGen solves these problems by using two main parts. One part is a graph transformer encoder, which learns how amino acids, the building blocks of proteins, interact with each other in a 3D space. It updates their positions to match how they would act in real life. The other part is a protein language model, which helps predict the sequence of amino acids. This model gets fine-tuned by PocketGen to make sure the sequence fits well with the protein’s shape.
New paths in bioengineering
The researchers have described PocketGen in a paper published in Nature Machine Intelligence.
Tests show PocketGen works better than older methods. It’s faster by ten times and makes proteins that bind more effectively to molecules like fentanyl and ibuprofen. PocketGen also compared well against other top models like RFDiffusion and RFDiffusionAA.
The tool also offers insights into why proteins bind the way they do, which helps scientists understand and predict protein behavior better. This advancement not only speeds up drug design but also opens new paths in biology and engineering by showing how AI can improve our understanding of life at the molecular level.
Let us know your thoughts! Sign up for a Mindplex account now, join our Telegram, or follow us on Twitter.

.png)

.png)


.png)


0 Comments
0 thoughts on “Deep learning system for protein design”